A DDBJ-KEGG-PDBj workflow: from pathways to protein-protein interactions
- For the details of the project, see DDBJ-KEGG-PDBj.
Members
- Yasumasa Shigemoto (WABI/SABI; DDBJ, Japan)
- Akira Kinjo (PDBj, Japan)
- Soichi Ogishima (Tokyo Medical and Dental Univ, Japan)
- Masumi Itoh (Hokkaido Univ)
The objective
Given a KEGG pathway ID of a specific species,
- Identify homologous proteins of the pathway.
- Infer possible protein-protein interactions between components of the homologous pathways.
How?
- Input: KEGG pathway ID
- Get amino acid sequences of all the proteins involved in the given pathway. (KEGG API)
- For each protein sequence, run BLAST against the UniProt (SwissProt / !TrEMBL) database to identify its homologs. (WABI)
- Sort the BLAST results according to species. (a local program)
- At this point, we will have a phylogenetic profile.
- For each BLAST hit (a UniProt entry), run BLAST against PDB. (PDBj SeqNavi API).
- Find physical interactions between different components. (a local program)
- That is, if two (UniProt) sequences are homologous to different chains of the same PDB entry, they are predicted to be in physical contact.
- Output the phylogenetic profile and the predicted species-wise PPI.
An example
- KEGG pathway (TCA cycle in E. coli): path:eco00020
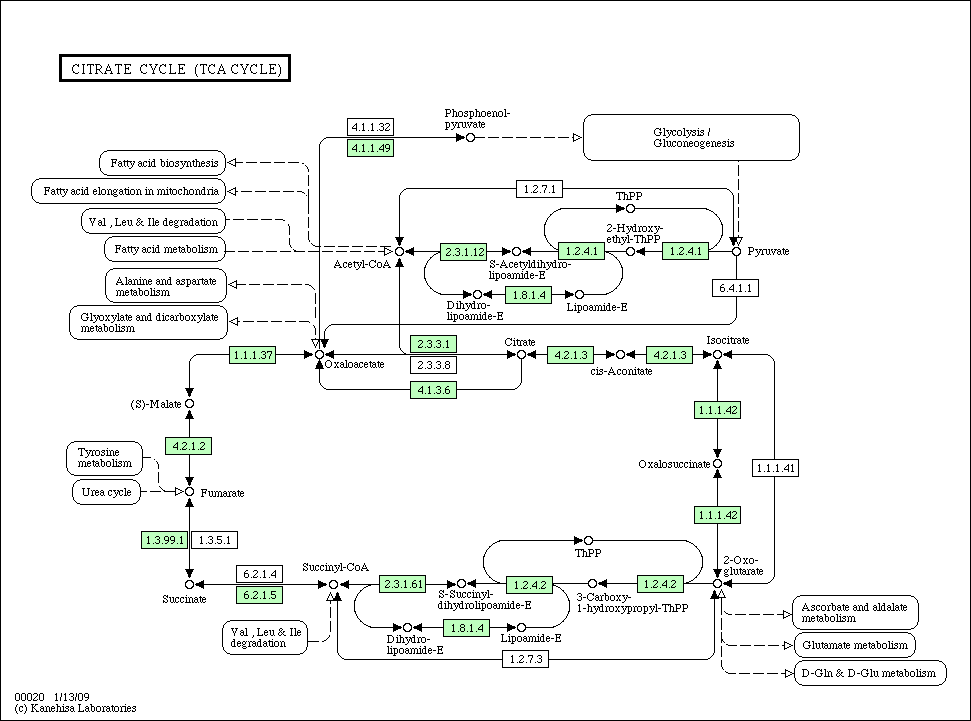 .
.
- Phylogenetic profile
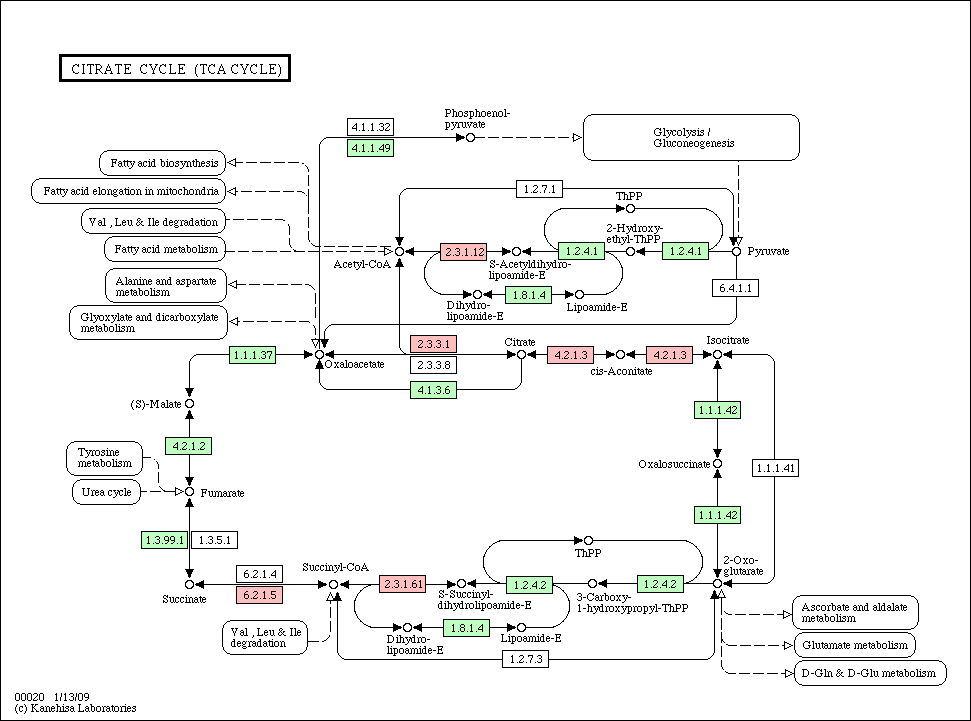
- The pathway colored according to homologs in Acinetobacter
- PPI list
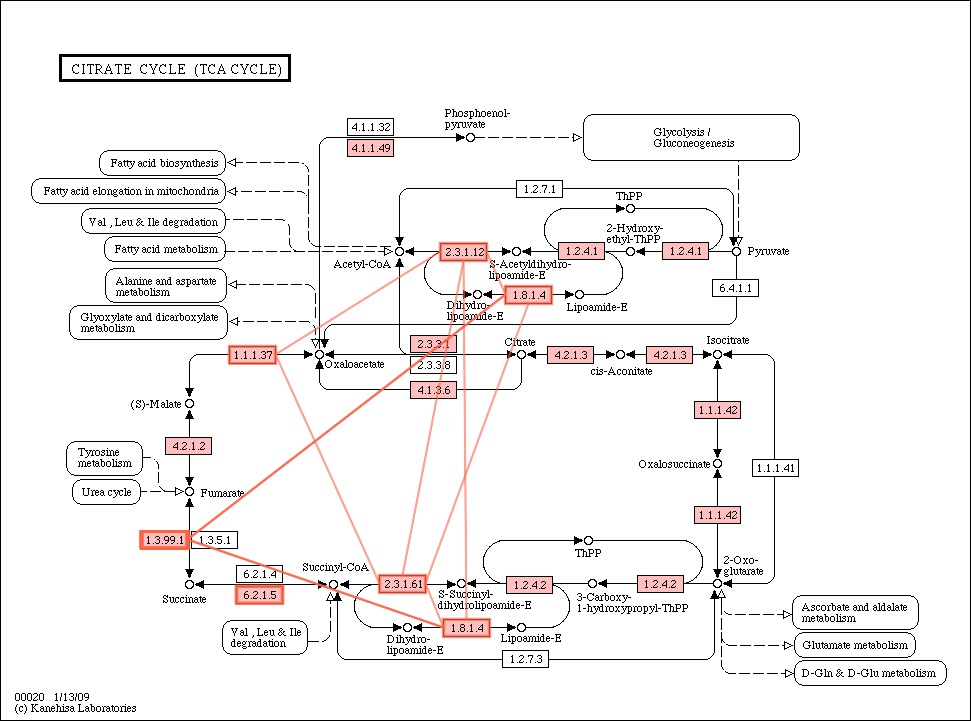
- The pathway with PPI edges in Yersinia pestis
What we learned
- We had to do a significant amount of coding in spite of the wealth of Web services.
- It takes a long time to finish all the flow.
- By actually solving biologically oriented problems, we can identify some typical use cases which might be useful for further development of web services.
- e.g. 1. Given a set of gene names, return a phylogenetic profile.
- e.g. 2. Given a set of blast hits, group them according to their species.
Attachments
-
sp_kegggene_matrix.2.xls
 (59.1 KB) - added by akinjo
16 years ago.
(59.1 KB) - added by akinjo
16 years ago.
a phylogenetic profile
-
Acinetobacter_sp._strain_ADP1_..gif
 (55.8 KB) - added by akinjo
16 years ago.
(55.8 KB) - added by akinjo
16 years ago.
pathway colored by phylogenetic profile
-
Yersinia_pestis.gif
 (25.1 KB) - added by akinjo
16 years ago.
(25.1 KB) - added by akinjo
16 years ago.
PPI
-
eco00020.gif
 (15.9 KB) - added by akinjo
16 years ago.
(15.9 KB) - added by akinjo
16 years ago.
eco00020
-
ppi_by_species.txt
 (80.3 KB) - added by akinjo
16 years ago.
(80.3 KB) - added by akinjo
16 years ago.
ppi list
