Satellite meeting for GlycoBiology?
Topics
Create and discuss about Taverna wolkflows for GycoBiology?.
Keywords:
- GlycoEpitopeDB http://www.glyco.is.ritsumei.ac.jp/epitope/
- H-InvDB http://h-invitational.jp/
- RINGS http://rings.t.soka.ac.jp/
- Consortium for Functional Glycomics (CFG) http://www.functionalglycomics.org
- GlycomeDB http://www.glycome-db.org
- (GlycoGeneDB) http://riodb.ibase.aist.go.jp/rcmg/ggdb/
- relations to diseases and SNPs
Chairperson
- Kiyoko F. Aoki-Kinoshita
Members
- Shujiro Okuda
- Shin Kawano
- Chisato Yamasaki
Targets
- Develop and register web services on BioMoby? such that glycobiologists can create workflows for glycobiology research.
- GlycoEpitopeDB:
- Query GlycoEpitope? DB using a query and retrieve all IDs of entries containing the keyword.
- Retrieve glycan structures in IUPAC format using GlycoEpitope? IDs.
- RINGS:
- Convert a glycan structure in IUPAC format into KCF format, by which glycan structure queries can be made.
- GlycoEpitopeDB:
- Create a workflow for analyzing glyco-gene-related diseases: Use OMIM to search for diseases related to loci of human homologs of target genes from other species, such as fruit fly. Retrieve any SNPs that are known to be related.
Summary
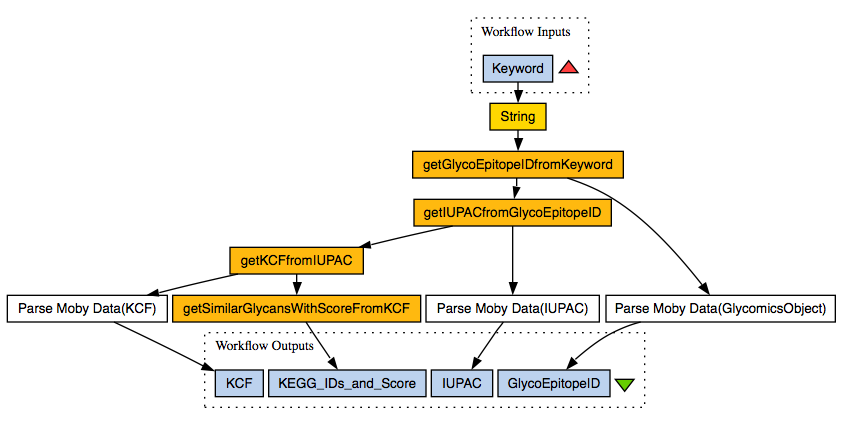
It took longer than expected, but we were able to create a BioMoby? web service in GlycoEpitopeDB.
- GlycoEpitopeDB
- getGlycoEpitopeIDfromKeyword
- getIUPACfromGlycoEpitopeID
- RINGS
- getKCFfromIUPAC
Our workflow thus takes as input a keyword and searches GlycoEpitopeDB for entries matching the keyword. The retrieved entries are then used to get the glycan structures from each entry in IUPAC format. A new utility in RINGS was then developed to convert from IUPAC to KCF format. In an additional step, we retrieved similar glycan structures to the converted ones in KCF format. However, we still need to develop a parser to retrieve the KEGG GLYCAN IDs along with their scores from the GlycomicsObject?.
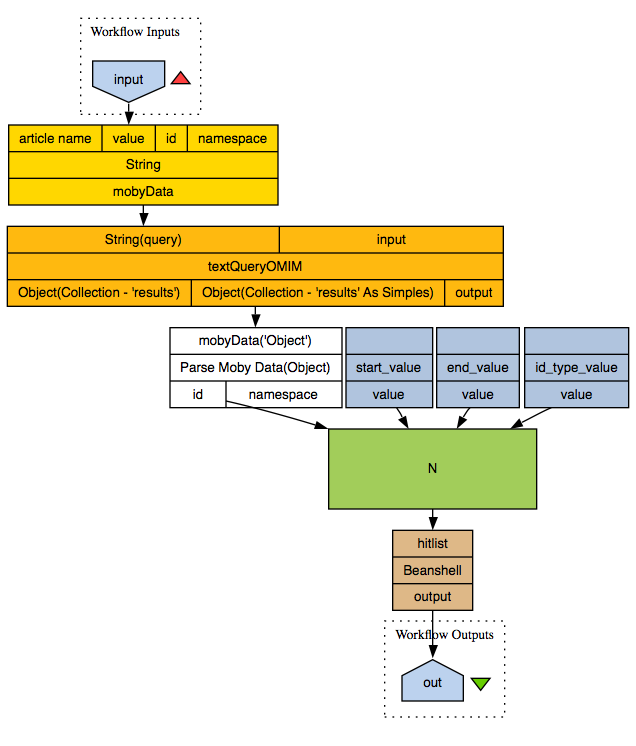
The disease-related workflow was able to retrieve H-Inv entries containing the OMIM Ids which were retrieved from OMIM for entries containing a particular keyword. The issue is that the output is in XML and contains entries which do not contain any SNPs, which we want to filter out. Tom showed us how to make a beanshell script which can be added to the workflow, and it worked!
Issues/Wishlist
- The Taverna 2 workbench could use some tweaking:
- When Taverna first starts up, there's a nice tree of all the web service activities that can be performed. However, after performing a search, there's no way to get that tree back.
- Some inconsistencies when using a Mac. No right-mouse button - it's usually mapped to ctrl-click, but it doesn't work all the time.
- BioMoby? installation with Perl-MoSeS is quite cumbersome.
- Apparently the cache on the server side of the web service needs to be updated in order for the BioMoby? web service to work. Intuitively, it doesn't seem necessary - as long as the web service code is on the server, it should work! This was one of the bottlenecks this week.
- It'd be nice to have a simpler way to install the code on the server.
- This may be a more general issue - I wasn't involved in the semantic web discussions at all, although I was interested. It'd be nice to have some hierarchical organization to the listing of web services in both BioMoby? and Taverna. Perhaps a semantically-oriented hierarchy?
Room
Select one:
- Chura Hall (3F)
Attachments
-
disease2snp.t2flow
 (26.5 KB) - added by kawano
16 years ago.
(26.5 KB) - added by kawano
16 years ago.
-
disease2snp.png
 (50.2 KB) - added by kawano
16 years ago.
(50.2 KB) - added by kawano
16 years ago.
-
glycoepitope_rings_wf.t2flow
 (27.4 KB) - added by shuo50
16 years ago.
(27.4 KB) - added by shuo50
16 years ago.
-
glycoepitope_rings_wf.png
 (50.2 KB) - added by shuo50
16 years ago.
(50.2 KB) - added by shuo50
16 years ago.